Computation
R Package lefko3
Package lefko3 is designed to provide a complete working environment to create and analyze matrix projection models (MPMs), including Leslie MPMs, Lefkovitch MPMs, historical stage-classified MPMs, integral projection models (IPMs), and age-by-stage MPMs, in both raw and function-based formats. It handles the entire workflow in MPM development, from formatting the demographic dataset, to building the life history model, to parameterizing vital rate models, to contructing the MPM, and finally to projection and analysis.
Package lefko3 works on R 3.6.3 or greater on all systems that support R, including Windows, MacOS, Linux, and Unix.
The project is hosted on CRAN and R-Forge. The most stable version of lefko3 is available for download through CRAN, and can be automatically downloaded with the install.packages() function in base R. The most recent development version is available through r-universe here. Documentation is available here. Please also see our paper and cite our paper describing lefko3 in Methods in Ecology and Evolution.
Current stable version: 6.4.0 (Mar 2025). Available on CRAN.
New in this stable version: Function supplemental() now handles additive offsets to matrix elements. All matrix creation and projection functions are able to handle these offsets.
Current development version: 6.4.0.9007 (Apr 2025). Available on r-universe.
New in this development version: Fixed bug resulting in fatal errors in age-by-stage MPMs with maximum ages smaller than those in the dataset. Function miniMod() now correctly handles zero-inflation models. deVries-formatted hMPMs are now indexed properly. Function modelsearch() no longer fails on historical runs with 2 vital rate variable entries.
Next stable version: 6.4.1 (mid 2025).
On the drawing board: Individual resampling functions. Fast linear model search function. Bootstrapping functions for vertical datasets. Adaptive dynamics projection and pairwise invasion analysis. Tests of reducibility and ergodicity. Complex IPMs with non-standard size functions. Demetrius entropy and Keyfitz entropy estimation.
Users of lefko3 may report issues with the package on Github. A Discussion community also exists within the lefko3 Github repository.
lefko3: a gentle introduction: a free online book teaching the use of lefko3. The official html version is available through Bookdown on R Studio Connect, at this site. To go to specific chapter, use the following links:
- Title, Copyright, and Preface The Basics
- Chapter 1: Introduction to R and lefko3
- Chapter 2: Preliminaries I: Life History Models
- Chapter 3: Preliminaries II: Data Formatting Matrix Models
- Chapter 4: Matrix Models I: Intro to MPMs, and Raw (Empirical) MPMs
- Chapter 5: Matrix Models II: Function-based MPMs
- Chapter 6: Matrix Models III: Age (Leslie), Hybrid Age, and Age-by-stage MPMs
- Chapter 7: Matrix Models IV: Integral Projection Models Projection Analyses
- Chapter 8: Population Projection I: Projection Simulations
- Chapter 9: Population Projection II: Deterministic Analysis
- Chapter 10: Population Projection III: Temporal Environmental Stochasticity
- Chapter 11: Population Projection IV: Density Dependence Special Analyses
- Chapter 12: Special Analyses I: Life Table Response Experiments (LTRE)
- Chapter 13: Special Analyses II: Quasi-Extinction Analysis Further Issues
- Chapter 14: Further Issues I: Importing Matrices and MPMs
- Chapter 15: Further Issues II: Importing IPMs and Function-based MPMs
- Chapter 16: Further Issues III: Editing Matrices in MPMs
- Chapter 17: Further Issues IV: Quality Control Appendices
- Chapter 18: Appendix I: Full Analysis Code Examples
- Chapter 19: Appendix II: Background Code for Chapters
- References: Literature cited More chapters will be added over time (currently planned chapter topics include transient dynamics, demographic stochasticity, and adaptive dynamics)
Tutorial materials already packaged with lefko3:
- Tutorial on theory, methods, and concepts
- Tutorial example of raw historical and ahistorical MPM analysis using Lathyrus vernus
- Tutorial example of function-based historical and ahistorical MPM analysis using Cypripedium candidum
- Tutorial example of historical and ahistorical IPM analysis using Lathyrus vernus
- Tutorial example of ahistorical age x stage MPM analysis using Lathyrus vernus
- Tutorial example of deterministic and stochastic LTRE analysis, and MPM import, using the Anthyllis vulneraria example published in Davison et al. 2010
Please contact the lab for further details about workshops.
Installation notes: Package lefko3 is available in its stable format on CRAN and can be downloaded with all relevant binaries from there. The latest development version, including all binaries, are available on r-universe.
Some users may occasionally wish to download and install the package from source. Package lefko3 is unlike the typical R package in that it relies on core kernels written in C++ to power its functions. This means that installation from source generally requires the installation of some development tools in order for R to install from source properly. If you are a Linux user, then your version of R is already set up to install lefko3 from source. However, if you are a Mac or Windows user, then you will need to complete a few extra steps to install lefko3 successfully from source.
Users may occasionally have issues with R not automatically installing the required packages for lefko3 to function properly when installed. Please make sure to install the following packages manually, in that circumstance (install in the following order): Rcpp, RcppArmadillo, BH, Matrix, TMB, lme4, pscl, VGAM, glmmTMB, Rcompadre, popbio. We also recommend the packages pryr and BAMMtools, for use with our user manual lefko3: a gentle introduction.
Mac users: To set up your Mac to install lefko3 from source, please follow these steps:
- Download and install the latest Xcode from the Apple App Store. This is a very large program - over 10GB. It provides the compilers and interfaces that the Mac version of R relies on to compile code.
- Download and install Xcode Command Line Tools. To do so, please open the Mac terminal, and type "xcode-select --install". This may open a diaglog box asking you if you would like to install developer tools. If this happens, then please click "Yes" or "Install".
- Download and install the latest GNU Fortran Compiler, available here. Choose the most appropriate build given your architecture (pay special attention to the OS version, and to the type of microprocessor).
- Open the Mac terminal back up, and type "export PATH=$PATH:/usr/local/gfortran/bin". This command adds GNU Fortran to the command path, thus allowing R to use it.
- Congrats! You should now be able to install from source!
Windows users: To set up your PC to install lefko3 from source, please follow these steps:
- Download and install Rtools.
- Add Rtools to the command path. See the section on this in the Rtools download page.
Biology

Evolutionary loss of photosynthesis
We are analyzing long-term demographic data and parameterizing evolutionary models to understand why so many plant species have lost photosynthesis and evolved to parasitize mycorrhizal fungi. Our work is both demographic and comparative, invloving matrix projection, adaptive dynamics, and phylogenetic analysis to understand these phenomena.
- Shefferson, R.P., K. Shutoh, and K. Suetsugu. 2024. Life history costs drive the evolution of mycoheterotrophs: increased sprouting and flowering in a strongly mycoheterotrophic Pyrola species. Journal of Ecology 112:1287-1300.
- Shefferson, R.P., M. Roy, U. Puttsepp, and M.-A. Selosse. 2016. Demographic shifts related to mycoheterotrophy and their fitness impacts in two Cephalanthera species. Ecology 97:1452-1462.
- Shefferson, R.P., M.K. McCormick, D.F. Whigham, and J.P. O’Neill. 2011. Life history strategy in herbaceous perennials: inferring demographic patterns from aboveground dynamics of a primarily subterranean, myco-heterotrophic orchid. Oikos 120:1291-1300.
Eco-evolutionary impacts of individual history
The events that occur in an individual's life can affect that individual in the long-term, and this impact can extend to long-term influences on population dynamics. For example, one of the greatest long-term impacts of individual history on mortality in herbaceous plants can come from high growth between years. We are studying the ecological and evolutionary dimensiomns of these impacts, and creating tools to study them.
- Shefferson, R.P., S. Kurokawa, and J. Ehrlén. 2021. lefko3: analyzing individual history through size-classified matrix population models. Methods in Ecology and Evolution 12:378-382.
- Shefferson, R.P., T. Kull, M.J. Hutchings, K.M. Kellett, E.S. Menges, R.B. Primack, K. Alahuhta, S. Hurskainen, H.M. Alexander, D.S. Anderson, R. Brys, E. Brzosko, S. Dostálik, K. Gregg, Z. Ipser, H. Jacquemyn, A. Jäkäläniemi, J. Jersáková, W.D. Kettle, M. McCormick, A. Mendoza, M.T. Miller, A. Moen, D.-I. Øien, Püttsepp, M. Roy, N. Sather, M.-A. Selosse, N. Sletvold, Z. Stipkova, K. Tali, J. Tuomi, R.J. Warren II, and D.F. Whigham. 2018. Drivers and characteristics of vegetative dormancy across herbaceous perennial plant species. Ecology Letters 21:724-733.
- Shefferson, R.P., M.K. McCormick, D.F. Whigham, and J.P. O’Neill. 2011. Life history strategy in herbaceous perennials: inferring demographic patterns from aboveground dynamics of a primarily subterranean, myco-heterotrophic orchid. Oikos 120:1291-1300.
Adaptive dynamics of sprouting
Why can so many photosynthetic plants live for years as rootstock with photosynthesizing? We address this question with a variety of evolutionary methods, including reconstructing trait evolution and game theoretical modeling. Our research has been featured in the news media, including on Nexus Media, Science Daily, or the Daily Mail.
- Shefferson, R.P., T. Kull, M.J. Hutchings, K.M. Kellett, E.S. Menges, R.B. Primack, K. Alahuhta, S. Hurskainen, H.M. Alexander, D.S. Anderson, R. Brys, E. Brzosko, S. Dostálik, K. Gregg, Z. Ipser, H. Jacquemyn, A. Jäkäläniemi, J. Jersáková, W.D. Kettle, M. McCormick, A. Mendoza, M.T. Miller, A. Moen, D.-I. Øien, Püttsepp, M. Roy, N. Sather, M.-A. Selosse, N. Sletvold, Z. Stipkova, K. Tali, J. Tuomi, R.J. Warren II, and D.F. Whigham. 2018. Drivers and characteristics of vegetative dormancy across herbaceous perennial plant species. Ecology Letters 21:724-733.
- Shefferson, R.P., R.J. Warren II, and H.R. Pulliam. 2014. Life history costs make perfect sprouting maladaptive in two herbaceous perennials. Journal of Ecology 102:1318-1328. (In the news)
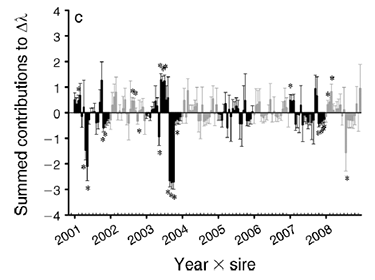
Drivers of plant demography
Why do plant populations vary in demographic response to climatic variation? How does genetic composition interact with environment to determine population dynamics? We work with herbaceous perennials to answer these questions.
- Shefferson, R.P., T. Kull, M.J. Hutchings, K.M. Kellett, E.S. Menges, R.B. Primack, K. Alahuhta, S. Hurskainen, H.M. Alexander, D.S. Anderson, R. Brys, E. Brzosko, S. Dostálik, K. Gregg, Z. Ipser, H. Jacquemyn, A. Jäkäläniemi, J. Jersáková, W.D. Kettle, M. McCormick, A. Mendoza, M.T. Miller, A. Moen, D.-I. Øien, Püttsepp, M. Roy, N. Sather, M.-A. Selosse, N. Sletvold, Z. Stipkova, K. Tali, J. Tuomi, R.J. Warren II, and D.F. Whigham. 2018. Drivers and characteristics of vegetative dormancy across herbaceous perennial plant species. Ecology Letters 21:724-733.
- Shefferson, R.P. and D.A. Roach. 2012. The triple helix of Plantago lanceolata: genetics and the environment interact to determine population dynamics. Ecology 93:793-802.
- Brys, R., R.P. Shefferson, and H. Jacquemyn. 2011. Impact of intrinsic and extrinsic variables on flowering and reproductive allocation patterns in a perennial iteroparous grassland herb: a ten-year experiment. Oecologia 166:293-303.
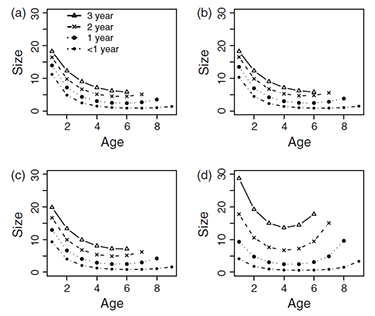
Plant and fungal senescence
We generally believe all organisms will senesce if they live long enough, but empirical assessments and some theory suggest otherwise. What determines how senescence evolves across the Tree of Life?
- Salguero-Gómez, R., R.P. Shefferson, and M.J. Hutchings. 2013. Plants do not count… or do they? New perspectives on the universality of senescence. Journal of Ecology 101:545-554.
- Shefferson, R.P. and D.A. Roach. 2013. Longitudinal analysis in Plantago: strength of selection and reverse age analysis reveal age-indeterminate senescence. Journal of Ecology 101:577-584.
