The Shefferson lab is devoted to computational evolutionary demography, with expertise in adaptive dynamics, matrix projection modeling, population dynamics, and life history evolution. We author and maintain the R package lefko3, a general programming environment for all matrix projection modeling, including integral projection, historical, age-by-stage, Leslie, and Lefkovitch models, with and without density dependence, environmental and individual covariates.
Our latest work

Why are some plants non-photosynthetic?
Some plant species shun the sunlight, instead relying on their soil-fungal symbionts for energy. Here, we compare fitness in sister species of Pyrola with different levels of mycoheterotrophy, finding increased sprouting with lessened reliance on photosynthesis.

Computing methods for MPM and IPM construction and analysis
This introductory manual shows how to construct and analyze matrix projection models and discretized integral projection models using R, with the complete MPM projection environment offered by R package lefko3.
Individual history and population dynamics
Ecologists typically analyze population dynamics ignoring individual history, but evidence suggests that history is important. Our R package lefko3 creates and analyzes matrix population models (MPMs) incorporating individual history.

Orchid life histories and demography
Orchids are prized for their unique and varied flowers. They have evolved special, long-lived life history strategies involving the production of dust seeds and dependence on fungal nutrition at least in the earliest stages of life. What other characteristics define their demography?
How do generalized interactions evolve?
Most of our understanding of coevolution comes from studies of specialist species. The complexity of generalist interactions has led too few studies of their coevolution. However, generalists must engage with partners that meet their ecological needs, and this must lead to an evolutionary pattern. How do these species coevolve?
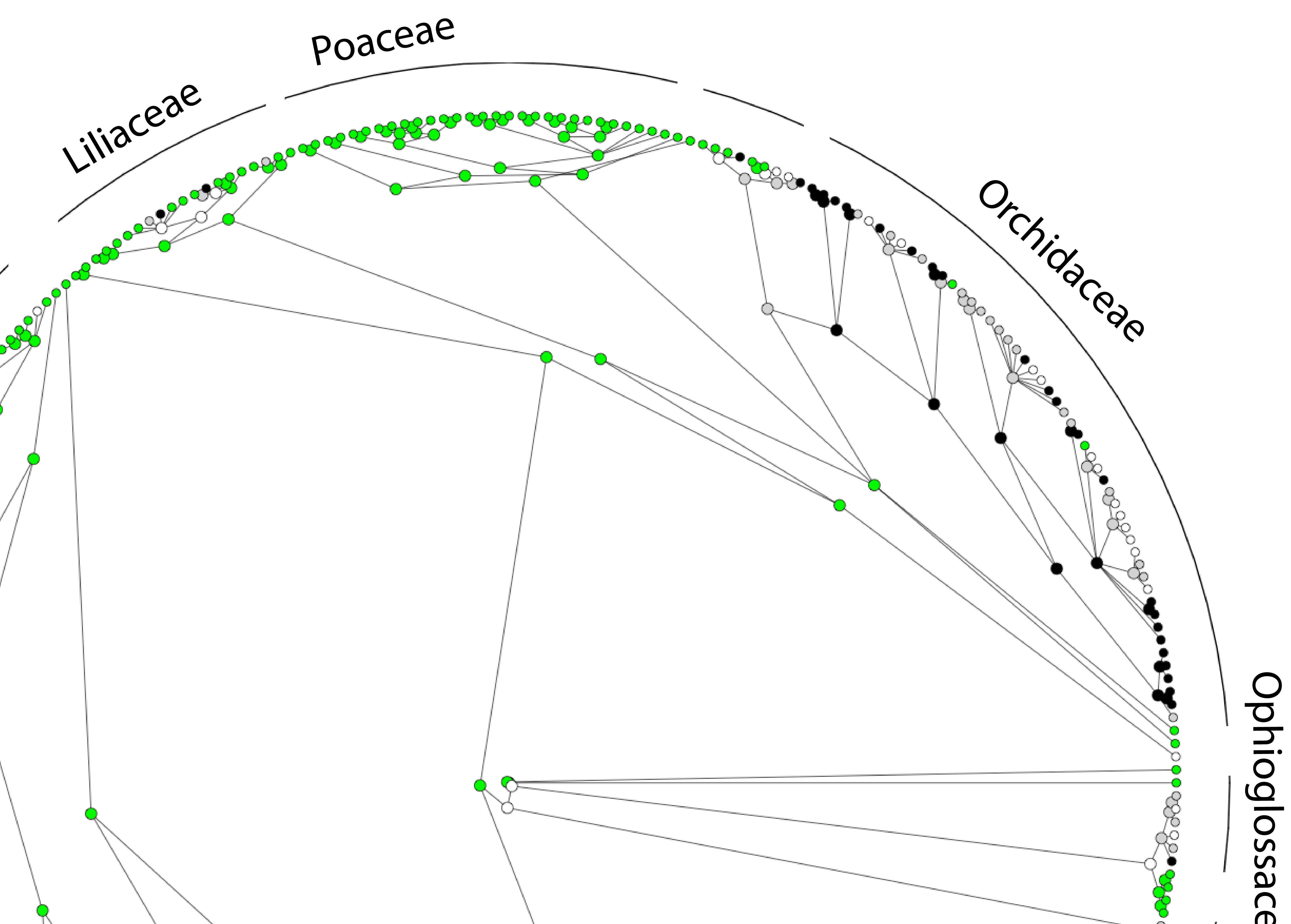
What drives vegetative dormancy?
Vegetative dormancy has been found in over 100 species and 24 families of plants, but what causes it, and what are its evolutionary origins? We led an international team to answer these questions. Read news articles about this research on Nexus Media, Science Daily, or the Daily Mail.

Can we predict evolution?
Contemporary climate change may cause many extinctions in the near future, but it is also likely to lead to rapid evolution. Can we predict how this evolution will occur? We believe so, and show how using vegetative dormancy in 3 orchid species.
In the blogs
- Apr 2024: Read our piece on the evolution of mycoheterotrophy in Pyrola species, on the Journal of Ecology Blog.
- Jun 2022: The Shefferson lab take on life as an ecologist in Japan.
- Dec 2020: Read our introduction to R package lefko3 on the Methods in Ecology and Evolution blog.
- Nov 2020: Read R. Shefferson's piece on the life histories and population dynamics of terrestrial orchids.
- Apr 2018: Read R. Shefferson's Ecological Inspirations piece in Journal of Ecology, about the paper that inspired him to become an evolutionary ecologist.
- June 2016: A commentary on a wonderful piece on latitudinal gradients in the Plant Kingdom by Zhang et al and in Journal of Ecology.
- July 2014: Read a commentary on a Journal of Ecology article dealing with why Rip van Winkle plants go dormant, particularly focused on life history costs.
